Making and Deploying an AI Web App in 2023 (Part 2)
Make a Proof of Concept for an AI App
This is part of a multi-part blogpost about how to build an AI Web App. Please refer to Part 1 for more context.
This post uses txtai as the AI library.
Alternatives would be: transformers, sentence-transformers, PyTorch, Keras, scikit-learn, among many others.
The first step in any project is always to make a proof of concept. At this stage, we don’t care about performance, edge cases, or any other intricacies— we just want to confirm that the project is viable.
For the sake of example, we will take strong inspiration from this example from the txtai library.
We’ll index a database of documents and then query for them with natural language.
Basically we’ll be implementing our own little Google Search, which only returns results from the files we feed it.
But we won’t just search for keywords, like classical search engines. It’s 2023 and we have AI at our disposal! We will use text embeddings to search for the closest match. By searching for embeddings we’re not literally searching for the words we give it, but for the meaning of the whole query. This is called Semantic search.
So, let’s get thing going by creating a new directory for our little project:
mkdir ai-web-app
cd ai-web-app
We will need a database to index and query.
For this example, we will use a test dataset that txtai made available.
This data is a collection of research documents into COVID-19.
The original version of this dataset can be found on Kaggle.
We download the dataset with the following shell commands:
wget https://github.com/neuml/txtai/releases/download/v1.1.0/tests.gz
gunzip tests.gz
mv tests articles.sqlite
and install the needed libraries with
pip install txtai sentence-transformers pandas jupyterlab
We can then start a notebook server with Jupyter Lab with
mkdir notebooks && cd notebooks && jupyter lab
From our database, we will need to create an index (a searchable database of
documents), and a way to query (search) the index.
The following code (mostly stolen from txtai’s example)
creates an index with the COVID-19 dataset downloaded above:
import sqlite3
import regex as re
from txtai.embeddings import Embeddings
from txtai.pipeline import Tokenizer
def stream():
# Connection to database file
db = sqlite3.connect("../articles.sqlite")
cur = db.cursor()
# Select tagged sentences without a NLP label. NLP labels are set for non-informative sentences.
cur.execute("SELECT Id, Name, Text FROM sections WHERE (labels is null or labels NOT IN ('FRAGMENT', 'QUESTION')) AND tags is not null")
count = 0
for row in cur:
# Unpack row
uid, name, text = row
# Only process certain document sections
if not name or not re.search(r"background|(?<!.*?results.*?)discussion|introduction|reference", name.lower()):
# Tokenize text
tokens = Tokenizer.tokenize(text)
document = (uid, tokens, None)
count += 1
if count % 1000 == 0:
print("Streamed %d documents" % (count), end="\r")
# Skip documents with no tokens parsed
if tokens:
yield document
print("Iterated over %d total rows" % (count))
# Free database resources
db.close()
# BM25 + fastText vectors
embeddings = Embeddings({
"method": "sentence-transformers",
"path": "all-MiniLM-L6-v2",
"scoring": "bm25"
})
embeddings.score(stream())
embeddings.index(stream())
In the code above, the text embeddings are generated using the all-MiniLM-L6-v2 model.
We could also train our own model instead of using a pre-trained model, but that
would require a lot of work.
If we can avoid it, why not?
So at this point, we have a database and want to search it.
In the following cell, we define a Python function to query the index for the closest results, and then query the original data for extra information about the results.
The last thing we do in this cell is to search for "risk factors".
import pandas as pd
pd.set_option("display.max_colwidth", None)
def search(query: str, topn: int = 5) -> pd.DataFrame:
db = sqlite3.connect("../articles.sqlite")
cur = db.cursor()
results = []
for uid, score in embeddings.search(query, topn):
cur.execute("SELECT article, text FROM sections WHERE id = ?", [uid])
uid, text = cur.fetchone()
cur.execute("SELECT Title, Published, Reference from articles where id = ?", [uid])
results.append(cur.fetchone() + (text,))
db.close()
df = pd.DataFrame(results, columns=["Title", "Published", "Reference", "Match"])
return df
search("risk factors")
The output of that cell will be a pandas dataframe with the results: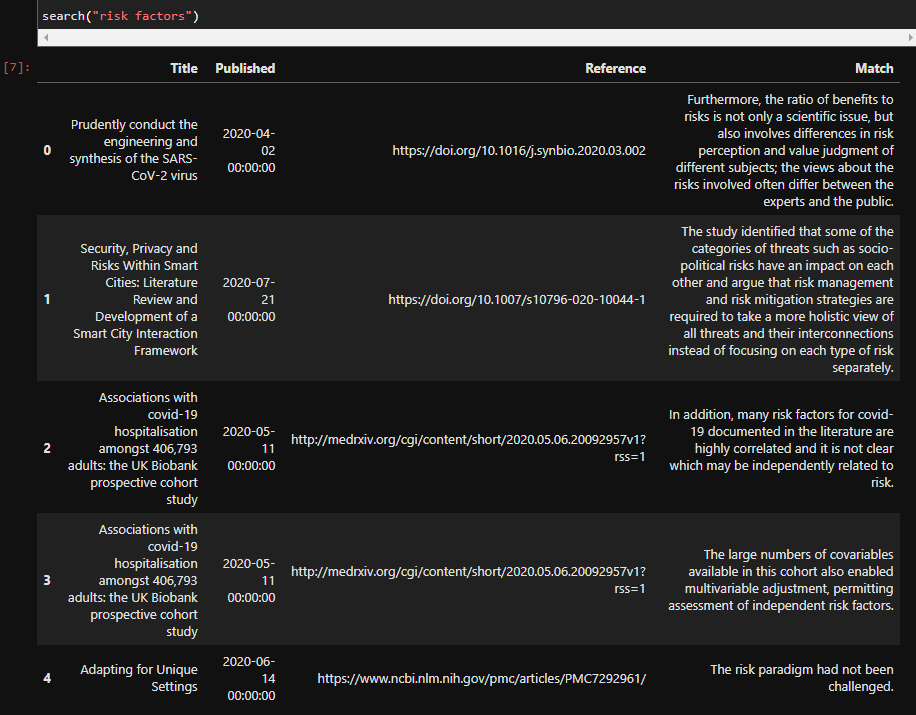
We can also try other queries: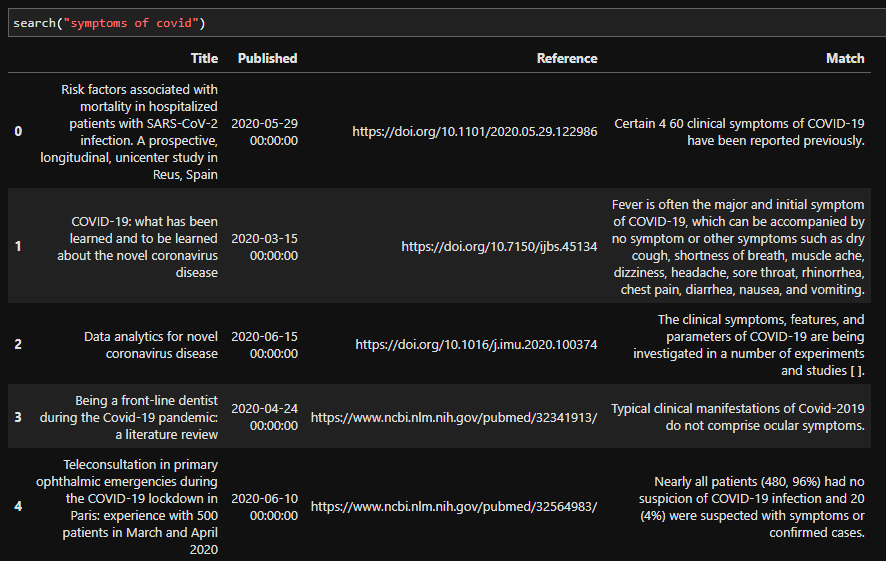
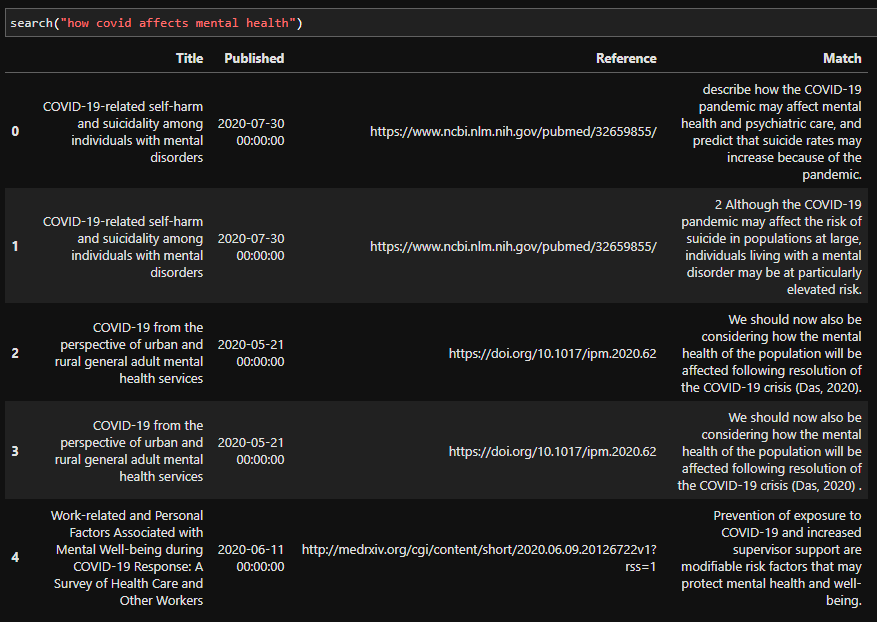
As you can see, while the results maybe aren’t the best, they are good enough for our proof of concept. We could spend a lot more time making this part better, but in the spirit of shipping early and often, this is enough for now.
To continue this tutorial, go to Part 3.
For comments or questions, use the Reddit discussion or reach out to me directly via email.